Effects of random forest modeling decisions on biogeochemical time series predictions
Abstract
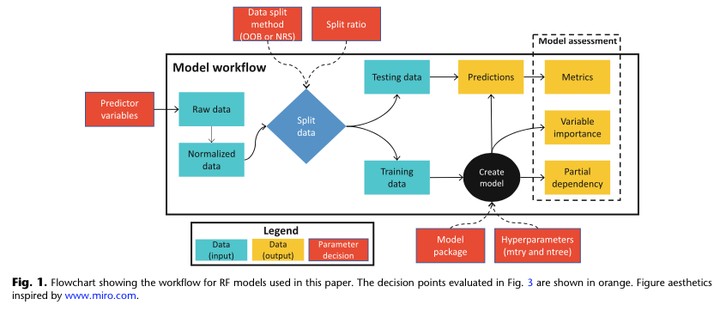
Random forests (RF) are an increasingly popular machine learning approach used to model biogeochemical processes in the Earth system. While RF models are robust to many assumptions that complicate deterministic models, there are several important parameterization decisions for appropriate use and optimal model fit. We explored the role that parameter decisions, including training/testing data splitting strategies, variable selection, and hyperparameters play on RF goodness-of-fit by constructing models using 1296 unique parameter combina- tions to predict concentrations of nitrate, a key nutrient for biogeochemical cycling in aquatic ecosystems. Models were built on long-term, publicly available water quality and meteorology time series collected by the National Estuarine Research Reserve monitoring network for two contrasting ecosystems representing freshwater and brackish estuaries. We found that accounting for temporal dependence when splitting data into training and testing subsets was key for avoiding over-estimation of model predictive power. In addition, variable selec- tion, the ratio of training to testing data, and to a lesser degree, variables per split and number of trees, were sig- nificant parameters for optimizing RF goodness-of-fit. We also explored how model parameter decisions influenced interpretation of the relative importance of predictors to the model, and model predictor-dependent variable relationships, with results suggesting that both data structure and model parameterization influence these factors. Because much of the current RF literature is written for the computational and statistical science communities, the primary goal of this study is to provide guidelines for aquatic scientists new to machine learn- ing to apply RF techniques appropriately to aquatic biogeochemical datasets.